A novel blood-based platform for monitoring liver disease
The oLIVER project team used a new method known as APTASHAPE to take 'pictures' of blood samples. Here you can read how the APTASHAPE method can be used to discover biomarkers and drug targets in liver disease.
He compares it to taking a picture. A picture of all the microscopic components in your blood. “The composition of your blood gives a robust indication of your state of health. When we take a picture of a blood sample and compare it to a control picture, from what we know is a healthy person, the differences between the two pictures can tell clinicians whether something might be wrong,” says Jørgen Kjems.
APTASHAPE – taking ‘fingerprint pictures’ of biofluids with RNA biosensors
Although Jørgen Kjems calls it a ‘picture’, there are no traditional cameras involved. Instead, he is using a molecular method called APTASHAPE to generate digital pictures composed of thousands of colored squares. “Each square represents the signal from one of the many millions of biosensors used in the APTASHAPE method. The biosensor are short RNA strands known as aptamers, which can bind to proteins in the blood sample. When a biosensor binds a protein, a signal is received,” Jørgen Kjems explains.
It sounds very simple, when put like this. In the lab, however, it’s a little more complicated. True enough, the basis of the APTASHAPE method is billions of different RNA biosensors and some of them bind to proteins in the blood sample. But the method also involves intermediate purification and sequencing steps as well as bioinformatical analyses. “When we have identified the biosensors that bind proteins in the sample, we separate them from the non-binders. The biosensors are then detached from the proteins and this is when the RNA nature of the biosensors come in handy,” says Jørgen Kjems and elaborates further: “Because the sensors are strings of RNA, modern sequencing techniques allow us to easily identify and quantify them. This approach enables us to generate a fingerprint-like ‘picture’: We use bioinformatical approaches to make a large matrix in a computer program. Each square in the picture represents a biosensor and the color of the square corresponds to the abundance of this exact biosensor.” The output is similar to a very pixelated digital picture and these ‘fingerprint pictures’ can then be easily compared by computer programs. In this way, the oLIVER team can quickly identify which biosensor have changed.
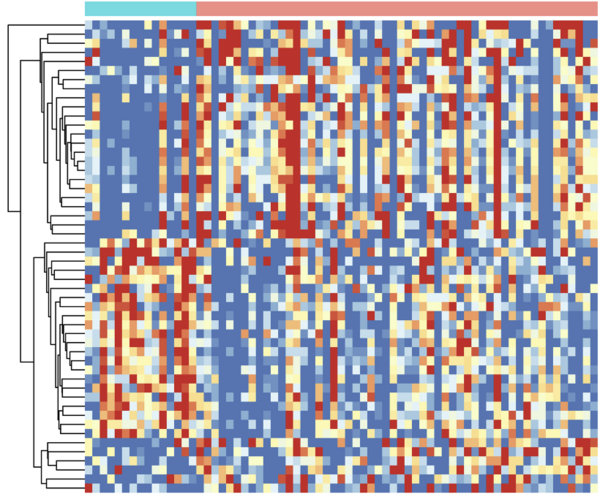
Using liver diseases as a case-study for testing the APTASHAPE method
The ability of the APTASHAPE method to generate a picture of blood samples – or basically any biofluid for that matter – was tested in the oLIVER project headed by Jørgen Kjems. As the project acronym implies, the project was an open collaboration on liver disease.
The project focused specifically on Metabolic Dysfunction-Associated SteatoHepatitis, or in short MASH. MASH is a complex metabolic disease where fat builds-up in the patients’ liver cells. In itself, the build-up of fat is harmless, but the liver tissue also becomes inflamed and scarred. “When inflammation and scarring accompany the build-up of fat, it can cause permanent scarring of the liver tissue. This may result in liver failure and increased risk of liver cancer,” says Jørgen Kjems. MASH is an increasing burden for society and although there is no simple explanation for the development of the disease, there is a clear correlation with metabolic disorders such as obesity and diabetes. Therefore, as the number of obese and diabetic patients rises, so does the number of patients with MASH.
“Clinicians cannot make a reliable MASH diagnosis without a liver biopsy, which is an invasive procedure” Jørgen Kjems states. “In the oLIVER project, we aimed to identify blood biomarkers for MASH using the APTASHAPE method. We know that the disease involves cell to cell signaling and therefore it should be possible to find biomarkers in the patients’ blood,” he reasons. The oLIVER team aimed for the biomarkers to be used in both diagnosis, disease staging, and monitoring of responses to drugs.
Biosensors can distinguish between healthy and diseased samples
In a research setting, mice are often used as a model for MASH – also at Novo Nordisk, which participated as an industry partner in the oLIVER project. The team therefore used the APTASHAPE method to analyze both mouse and human blood plasma samples. Plasma is the fluid part of the blood, which transports the blood cells in our veins - and without the blood cells it is much easier to analyze the samples. The mouse samples were provided by Novo Nordisk, whereas the human samples were supplied from Aarhus University Hospital. “The human plasma samples came from well-characterized and biopsy-verified MASH patients at different stages of the disease. Because we also received healthy samples as a control, we set out to investigate whether APTASHAPE could distinguish MASH patients from healthy individuals and also between the early and late stages of MASH,” explains Jørgen Kjems.
Using the APTASHAPE method, Jørgen Kjems and his team identified several specific RNA biosensors that could predict MASH disease in blood plasma samples. The biosensors could also distinguish between different stages of MASH. “We saw a correlation between the APTASHAPE picture and the patient profile - just like we expected,” Jørgen Kjems says. He further explains that the APTASHAPE method was also used on mouse samples to monitor the treatment response of drugs. Here, mice were fed either a lean or a Western diet. The former mice were healthy controls, whereas the mice on a Western diet developed MASH. The mice suffering from MASH were treated with two different drug candidates or placebo. “In total, we had five different mouse groups. We used the APTASHAPE method to take ‘fingerprint pictures’ of the different groups’ blood samples and we saw a clear difference between the healthy control and the MASH mice. We could clearly observe an effect of the drugs,” says Jørgen Kjems and elaborates: “When we compared the APTASHAPE ‘fingerprints’ of the plasma samples, it was evident that the drug candidates pushed the APTASHPAE ‘fingerprint’ back towards the lean diet picture. We believe that this is an indication that the drug candidates are effective.”

Protein ‘fishing’ – identifying biomarkers using APTASHAPE
The APTASHAPE method also has other applications. One of them is to identify the proteins that change during disease. This is possible because each RNA biosensor binds a specific target: “It’s a bit like the old magnetic fish game for toddlers,” says Jørgen Kjems and explains the analogy: “When we had identified the biosensors that represented the biggest differences between healthy and diseased blood samples, we attached small magnetic beads to these specific biosensors. As soon as the magnetic biosensors had bound to the proteins, we used a magnet to ‘fish’ out the biosensors from the rest of the molecules in the blood samples – the proteins followed along the biosensors, which they had bound to.”
When the proteins had been ‘fished’ out of the plasma, the oLIVER team used a standard technique known as mass spectroscopy to identify the proteins that were either up- or downregulated in MASH. Using this ‘fishing’ strategy, the oLIVER team identified a list of proteins that can possibly serve as biomarkers in MASH. “We were happy to see that the proteins we identified also had been suggested as potential biomarkers in other studies,” says Jørgen Kjems. “In the oLIVER project, we have validated other research experiments, we have automated the method in collaboration with Novo Nordisk, and we have demonstrated the usefulness of the APTASHAPE method,” he says.
The oLIVER project serves as a stepping stone for future research and innovation
The oLIVER project is central to a range of Jørgen Kjems’ future plans, because it has demonstrated the applicability of the APTASHAPE method. “I have already planned – and also initiated – new collaborations with both new and former collaborators where we will use the APTASHAPE method. In this regard, the oLIVER project served as a stepping stone for many of these ideas,” he says. “It was a great opportunity for the oLIVER team to work in a close collaboration with Novo Nordisk in a project without IP. It is my impression that it has been very satisfying for both us in academia and for the industrial partners, because there were no economic ties between us and no lengthy negotiations over IP,” Jørgen Kjems elaborates.
Some of Jørgen Kjems’ many plans include academic collaborations within other disease areas such as cardiovascular disease, gastro-intestinal disease, leukemia, and kidney diseases. He is also hoping that the oLIVER project can form the basis for further collaborations with Novo Nordisk on liver diseases. At the same time, he is very enthusiastic about the interest from other companies: “I have been contacted by several companies specializing in MedTech devices. They have been eager to hear more about the possibility of implementing the APTASHAPE method in a quick test format – similar to the COVID-19 self-test assays used during the pandemic,” he says.
Although time will show what the future holds for the APTASHAPE method, there is no shortage of ideas from Jørgen Kjems’ side: “From my perspective, the combination of the APTASHAPE method and machine learning tools is very powerful. The more APTASHAPE ‘fingerprint pictures’ we have of blood samples from different diseases, the greater possibility we have to use machine learning tools to reveal yet unknown links between different diseases,” says Jørgen Kjems. “If we do this in an open setting without IP, the results can serve as a basis for accelerating knowledge and downstream innovation,” he concludes.
You can find more information about the oLIVER project here.
