Researchers at DANDRITE and collaborative partners have used cutting-edge electron microscopy to reveal the first structures of the P4-ATPase lipid flippase; a crucial protein for cell viability
PhD-student Milena Timcenko and Assistant Professor Joseph A. Lyons give an insight into their recently published article in the renowned journal Nature “Structure and autoregulation of a P4-ATPase lipid flippase”.
PhD-student Milena Timcenko has an MSc in Nanoscience. During her PhD, she has studied the structures of yeast lipid flippase Drs2p-Cdc50p and has expertise within cryo-EM data processing. Assistant Professor Joseph A. Lyons has a background in Chemistry. He transitioned from lipid chemistry to investigate membrane protein structure and function using in vogue advanced technologies in structural biology.
Can you tell me about the biological function of the P4-ATPase lipid flippase?
Milena explains: “All cells are surrounded by lipid-bilayer membranes. In eukaryotic membranes, the distribution of different lipids between the two sides of the membrane is highly specific and of great importance in multiple cellular processes. The role of the P4-ATPase is to regulate the distribution of phospholipids. They transport specific lipids from the extracellular leaflet to the intracellular leaflet against their concentration gradient; thereby restricting some lipids to just the inner leaflet. Joseph comments, “The lipid distribution between the two sides of the membrane is asymmetric. This means, these lipids can be used for signaling because you have an ‘on’ and ‘off’ switch. The on-switch is a family of proteins called lipid scramblases, which allow lipids to return to the outer leaflet of the membrane, while the P4-ATPase lipid flippase would be the off-switch”.
What does the paper reveal?
Our structures reveal the architecture of the P4-ATPase lipid flippases and three specific conformational states of our protein of interest, Drs2p-Cdc50p. We learn about how the autoinhibition works and how this P4-ATPase is autoregulated. Other P-type ATPases that we know are autoinhibited and may share similar properties. One of the big questions in the field of P4-ATPases has been, how lipids are transported compared to cations that are the substrate of significantly more studied P-type ATPases. The determined structures enable us to propose a possible mechanism for lipid transport.
What knowledge does the study contribute with in terms of cell biology, human health and disease?
In general, P4-ATPase flippases are essential proteins for cell viability. Mutations in these proteins have been linked to numerous diseases such as obesity, diabetes, Alzheimer’s and other neurodegenerative disorders. A structure is a fundamental step in trying to understand the protein and its function in a greater context. It is also a really good scaffold for interpreting available biochemical data. These proteins have an important role in cell biology by shaping the membrane environment, thus, influencing proteins and pathways reliant on this environment. All substances that influence the cell must interact with the membrane. If you have a membrane dysfunction in terms of loss of asymmetry or disrupted distribution of lipids, this basically alters the chemistry of the membrane and can be central to the pathogenesis of disease.
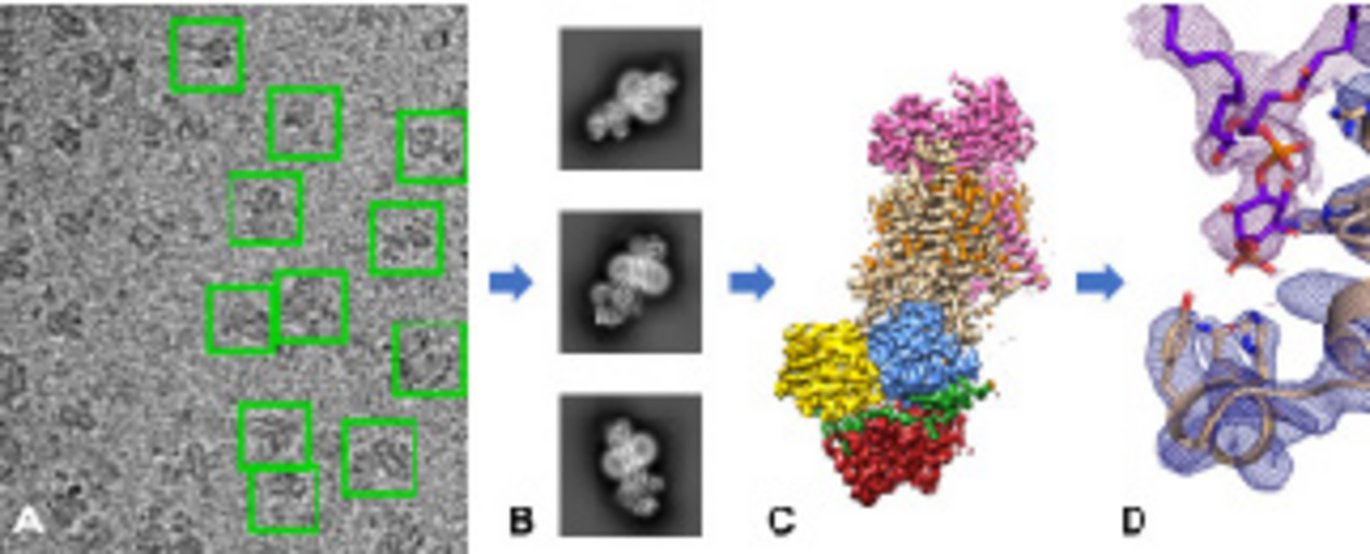
The structure of P4-ATPase is determined using cryo-electron microscopy. Can you tell me about the main advantages of using cryo-EM compared to e.g. X-ray crystallography?
Milena: “One of the major advantages of using cryo-EM is, that you do not need a crystal. Some proteins are difficult to crystallize and there are a number of chemical and physical parameters that need to be balanced for successful protein crystallization. In this case, cryo-EM has certainly been an advantage since scientists have spent many years trying to crystallize this protein without success”. Joseph continues: “Another advantage of using cryo-EM is that we can study protein conformational heterogeneity by sampling multiple conformational states from the same experiment. In crystallography you are typically restricted to one protein conformation locked within the crystal lattice so you would not get the ensemble information that is possible with cryo-EM. In our study, once we had collected the first dataset, a few months later we were able to collect two additional datasets giving us the structures in three different conformational states.”
What has been the most challenging aspect in this project?
This is the first protein structure solved by cryo-EM from our research group. It has been a learning curve for us along the way from sample preparation to using the microscopes and data processing. We attempted numerous strategies by following the literature, however they did not go as well as we had hoped. Going back to what we knew and trusted made the difference. We made a huge progress working out how to prepare a good cryo-EM grid which has played a great part in this project. Joseph concludes: “The challenge has been learning by doing”.
How does cryo-EM complement structural biology?
Cryo-EM is playing a huge role in structural biology, especially in the light of larger biological complexes and proteins that are difficult to crystallize. Cryo-EM is a very powerful tool that makes it possible to image not only large macromolecular complexes but now also small proteins at almost atomic resolution. The next big step for structural biology is to correlate information from a variety of structural determination methods to answer the bigger questions in life science: how does it all work together in a cell?
What will you be looking into next?
We want to put this protein in perspective and study these proteins in larger assemblies of membranes. Specifically, for Drs2p-Cdc50p there are other aspects of the lipid transport and autoregulation that we have not explored yet. In compliance with published data our structure suggests that the regulatory lipid we visualize activating the protein, is not the only factor that activates the protein. We observe the protein remaining in an autoinhibited or partially autoinhibited state. Therefore, the next goal is to look at the extra regulators that fully activate the protein. We can visualize that from a structural context by cryo-EM.
Further information:
- Link to the scientific article https://www.nature.com/articles/s41586-019-1344-7
-
Learn more about the cryo-EM facility at Aarhus University http://inano.au.dk/research/research-platforms/inano-cryo-em-facility/