Miniaturized version of ribosome found in microsporidia
A research team lead by MIMS/SciLifeLab research group leader Jonas Barandun uses cryo-electron microscopy to provide near atomic details of the smallest known eukaryotic cytoplasmic protein synthesis machine, the microsporidian ribosome.

150 years ago, the European silk industry was threatened by an unknown epidemic killing the silkworms. At that time, Louis Pasteur was able to identify the source of infection and made important suggestions for treatment. The silk production in Europe survived. Today, a microsporidian parasite is known as the cause of this epidemic and silk worm diseases still cause more than 100 million USD losses to the Chinese silk industry every year. Microsporidiosis is not restricted to silk worms. The diverse phylum of the microsporidia contains thousands of different species with parasites for essentially every animal. At least 14 of them can infect humans. Particularly challenged by microsporidia are not only aquacultures, sericultures and honey bee populations in which infections can wipe out entire hives, but also immunocompromised patients. Microsporidia are a risk for the environment, agriculture and human health and the US National Institutes of Health (NIH) recently added the parasitic fungi to the list of emerging pathogens of high priority. Even if microsporidia infections are among the most common parasitic diseases in all animals, relatively little is known about their fascinating molecular life which is shaped by an accelerated evolutionary rate and extreme genome compaction.
Together with researchers from The Rockefeller University and Connecticut Agricultural Experiment Station, Jonas Barandun, new group leader at The Laboratory for Molecular Infection Medicine Sweden (MIMS), publishes the cryo-electron microscopy structure of the microsporidian ribosome which visualizes the effect of extreme genome compaction on an essential molecular machine (Nature Microbiology, 22 July 2019).
The smallest eukaryotic genome
Microsporidian parasites can survive as spores in soil, water and air where they arrest in a dormant state. Once ingested by a host, they use a unique, ultra-fast infection mechanism to inject the entire content of the spore into the host cell. Once inside of a cell, microsporidia steal small molecules, such as ATP, from their host organism. This parasitic nature allowed them to lose many important genes to produce these small molecules. This compaction resulted in the smallest genome ever described in eukaryotes – even smaller than some bacterial genomes – containing approximately 2000 highly compacted genes.
Jonas Barandun commented “Microsporidia are the minimalists among the parasitic eukaryotes, reducing their genome to a minimum which is needed for survival and replication. This makes them ideal model organisms to study minimally required components of a molecular process”.
Unexpected findings with the help of cryo-electron microscopy
The first challenging step was to obtain sufficient cellular amounts to extract ribosomes. To overcome this challenge, Jonas Barandun teamed up with a microsporidia specialist, Charles Vossbrinck, from the Connecticut Agricultural Experiment Station, who grew the microsporidium Vairimorpha necatrix in larvae of the corn earworm Helicoverpa zea, a pest that can cause big damage to cotton and corn crops. After extraction of spores from the host organism, ribosomes were isolated from them and cryo-EM and mass-spectrometry studies were performed in the laboratory of Sebastian Klinge, a ribosome specialist at Rockefeller University in New York, USA, together with Mirjam Hunziker. This allowed the team to provide a near-atomic model of the smallest known eukaryotic cytoplasmic ribosome.
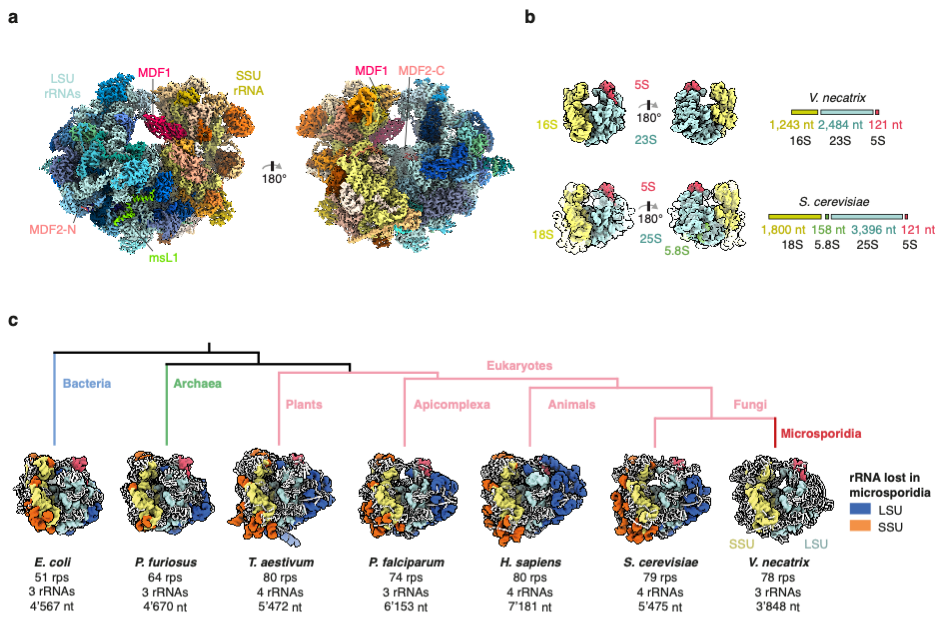
In the studied organism, Vairimorpha nectatrix, the ribosomal RNA is approximately 30% shorter than the rRNA in yeast and even 15% shorter than the bacterial rRNA in E. coli.
“While it was known that the microsporidian rRNA is significantly smaller than the related yeast rRNA, it was unclear if this compacted rRNA still was able to bind all the eukaryotic ribosomal proteins. Surprisingly, despite the loss of some of their RNA binding sites, almost all ribosomal proteins were still present in the structure, some of them exclusively bound by other ribosomal proteins and not in contact with RNA anymore” explains Jonas.
Compared to the closest related ribosome structure from fungi, microsporidian ribosomes have lost only two ribosomal proteins. In one region of the ribosome, where several RNA elements have been removed, a previously unknown microsporidia specific protein compensates for the extensive loss of RNA and serves as a placeholder.
During their evolution to organisms with highly compacted genomes, microsporidia have removed essentially all eukaryotic expansion segments – insertions present in the eukaryotic ribosomal RNA. The findings represent a reversion of the evolutionary expansion found in eukaryotic ribosomes. In eukaryotic ribosomes, expansion segments interact extensively with ribosomal proteins and a loss of these elements could also coincide with loss of the proteins that are bound to them.
“The most surprising finding was that the characterized ribosome appeared to be functionally inactivated by two microsporidian dormancy factors (MDF1, MDF2) and we can now assign a potential role to these two proteins of unknown function”, says Jonas.
Ribosomes were isolated from microsporidian spores, the extracellular dormant spore stage of the organism. As the parasite depends heavily on the resources of its host, an efficient shutdown mechanism for cellular processes could be advantageous to preserve energy during the spore stage. One of these two identified factors exist in all eukaryotic organisms, even in humans, but its role remained elusive. Future work will be required to confirm a similar role of this protein in other eukaryotic organisms.
- For more information visit http://www.mims.umu.se/groups/jonas-barandun.html
- For more information on Barandun lab visit https://barandunlab.org/
- Visit https://www.nature.com/articles/s41564-019-0514-6 to read the article published in Nature Microbiology ” Evolutionary compaction and adaptation visualized by the structure of the dormant microsporidian ribosome”.
- Contact information: Jonas Barandun PhD, group leader and SciLifeLab National Fellow, The Laboratory for Molecular Infection Medicine Sweden, MIMS, Umeå Centre for Microbial Research, UCMR, Department of Molecular Biology, Umeå University, Sweden Phone: +46 (0)70 717 95 11, Email: jonas.barandun@umu.se
Collaboration partners in this study:
Miriam Hunziker, PhD (now MIMS) Sebastian Klinge, PhD, Associate Professor, Laboratory of Protein and Nucleic Acid Chemistry, Klinge Lab, The Rockefeller University, New York, USA https://klingelab.rockefeller.edu
Charles R. Vossbrinck, Department of Environmental Sciences, The Connecticut Agricultural Experiment Station, New Haven, CT, USA https://portal.ct.gov/CAES/ABOUT-CAES/Staff-Biographies/Charles-R-Vossbrinck