Microbiome Pillar
Microbiome Pillar Overview
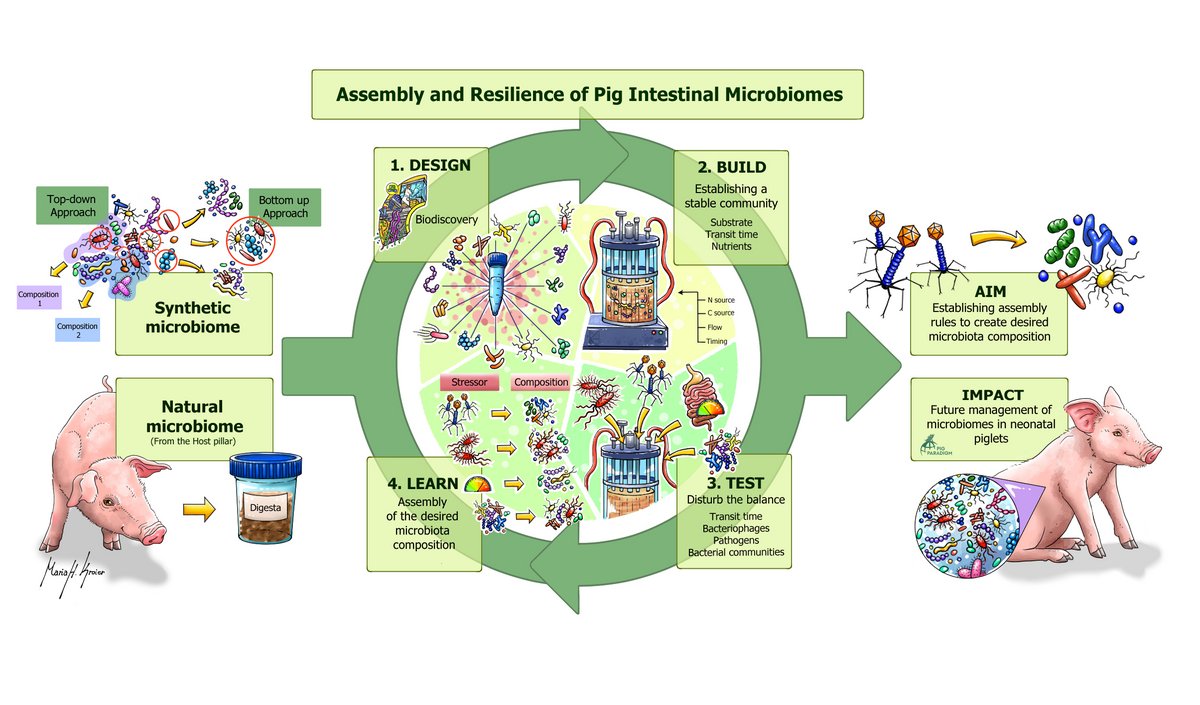
The Microbiome pillar is tightly aligned with the other pillars through integrating information from the large prospective phenotyping cohort (Host pillar) with data from small-scale controlled/intensive longitudinal in vivo studies (Nutrition Pillar) as well as mechanistic in vitro and in vivo experiments using natural as well as synthetic microbiomes, in vitro host models as well as a gnotobiotic mouse model, selected luminal metabolites and advanced stem cell based models of the host intestine. State-of-the-art bioinformatics will be employed that allow characterization of the antibiotic resistome as well as to provide strain-level resolution of microbial composition and functionality in complex microbiomes (Data Integration Pillar). In this highly integrative, multidisciplinary pillar, we will use hypothesis-driven and exploratory research to investigate the overarching hypothesis: Understanding of fundamental driving forces and manipulation of the intestinal microbiome can be a powerful tool to maximize resilience of the developing microbiome and its host, and thus minimize the need for antimicrobial therapy, and spread of AMR.
Research in the Microbiome Pillar has the following key focus areas:
- Establish the biogeography of the developing ‘healthy’ and ‘unhealthy’ intestinal microbiome during the first 12 weeks of life, with focus on all members of the intestinal microbiome, including bacteria, archaea, fungi, protozoa as well as viruses/phages
- Identify the key members and their functions that govern proper early assembly towards a resilient and robust intestinal ecosystem; establish representative (synthetic) microbiomes
- Establish driving forces of postweaning microbiome resilience and pathogen invasion, particularly in relation to post-weaning diarrhea
- Employ mechanistic in vitro co-culture and in vivo gnotobiotic animal models to gain mechanistic understanding of the developing host-microbiome interplay from birth and during the weaning transition, particularly at the mucosal host-microbiome interface
The hypothesis and focus areas will be investigated in the following Microbiome Research Themes (MRTs):
- MRT1: Biogeography of the developing intestinal microbiome
- MRT2: Design rules & driving forces of microbiome assembly and resilience
- MRT3: Mechanistic in vitro models of the developing host-microbiome interplay