From Samples to Knowledge
In an ever-changing world where the digital revolution and artificial intelligence are increasingly integrated into our daily lives, the question arises: how can modern technologies enhance the effectiveness, objectivity, and reproducibility of science?
Recently, Cecilie Brandt Becker, an Early Career Researcher in PIG-PARADIGM, participated in a workshop dedicated to building knowledge and skills in the utilization of computer software to transform tissue samples into valuable biological information.
Read this blog to gain insights into how these technologies are leading the conversion of traditional microscopy to digital image analysis and supporting Cecilie's work in PIG-PARADIGM.
By Cecilie Brandt Becker
With a background in veterinary medicine and special pathology (i.e. the study of disease processes in animals), my trainings have been largely rooted in the traditions of tissue analysis using a standard microscope. But in the ever-changing world, digitalization and the implementation of artificial intelligence is all around us, and the call for effectiveness, objectivity and reproducibility in science has never been bigger.
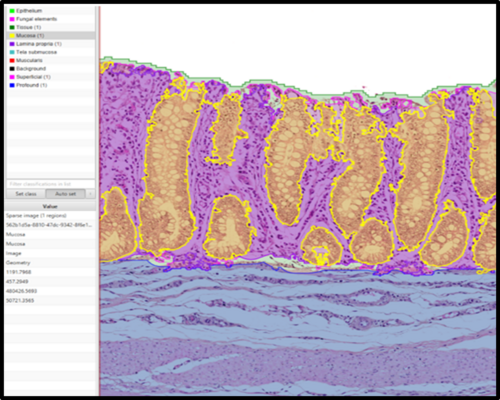
From a pathologist’s point of view, this change includes the conversion of traditional microscopy to digital image analysis, where computer software is trained to recognize cells and tissue patterns (inflammation, cancer, scar tissue formation etc.) in order to analyze tissue samples in a more standardized, effective and objective manner.
In the PIG-PARADIGM project, my research within the Host Pillar deals with the analysis of intestinal tissue samples from over 200 piglets. The main focus is on the inflammatory response and the interaction between the host (i.e. the pig) and the bacteria present in the intestinal content, and how these factors influence the pig’s resistance to development of diarrhea around the time of weaning.
To facilitate the analysis of this substantial volume of tissue samples, the implementation of digital image analysis seems like the obvious choice, and I will be using the free, open-source software QuPath. In order to improve my skills in this software, I recently attended an intensivetraining course hosted by the La Jolla Institute for Immunology in San Diego, CA, USA.
The course included a 3-day intensive training program in the approach to automated tissue analysis using QuPath, covering the basics in programming, deep learning models and application of a more quantitative approach to tissue analysis. The course was an amazing opportunity for me to gain some insight into the implementation of digital image analysis in relation to my Ph.D. project, and to update my toolbox of skills with regards to QuPath.
Moreover, it was an excellent experience, with substantial expansion of my scientific network that opened doors for possible future collaborations within my field of histopathology.