Decoding the competition pattern among rhizobia for the formulation of elite inoculants to enhance alfalfa production
The selection of rhizobia with high competition abilities, and more adapted to stressful conditions, is fundamental to formulate elite inoculants for enhancing alfalfa yield and improving its growth in marginal areas.
In the race to develop improved rhizobial inoculants, screening for rhizobia with high nitrogen fixation efficiency may not be the sole criterion for field application. Indeed, the selected rhizobia have to overcome different adverse conditions in soil (as salinity and drought) and outcompete other indigenous rhizobial strains. In this context, the activities of the first 9 months of the ALL-IN project were related to the development of elite inoculants to boost alfalfa production in harsh conditions based on two main phenotypes: high competition capabilities and resistance to dry environments. With this purpose:
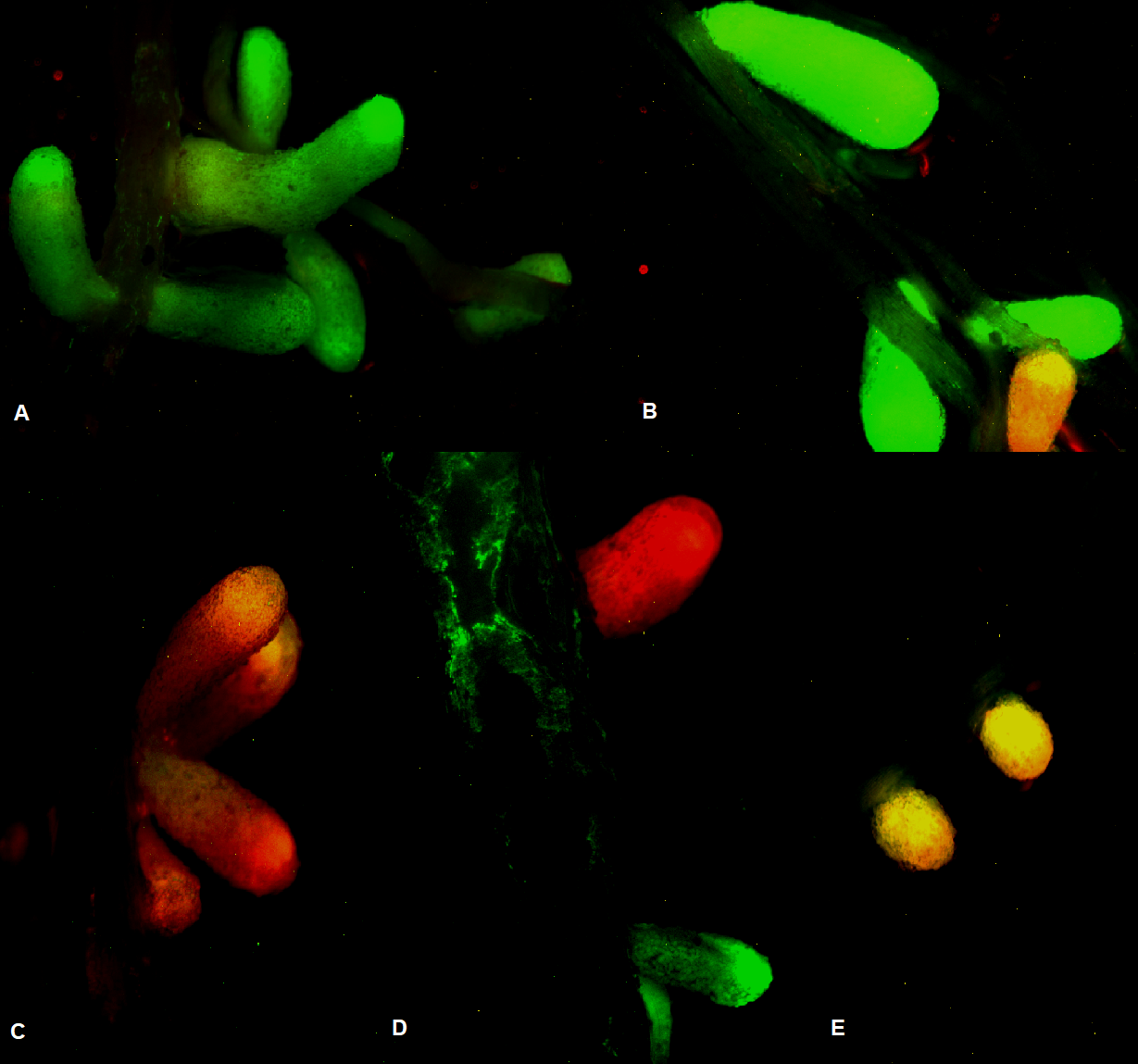
- The genomic determinants able to increase competition capability in the model rhizobial species Sinorhizobium meliloti were discovered. Competition tests were performed inoculating alfalfa seedlings with a rhizobial mix of two different fluorescent-labeled strains at a time. In particular, 13 S. meliloti strains GFP-tagged were tested against three well-characterized reference strains RFP-tagged (Rm1021, BL225C, and AK83), and the nodule occupancy was assessed using an epifluorescence stereomicroscope (Photo 1). To define the genetic features responsible for the competition phenotype, previously assessed in our tests, a genome-wide association (GWAS) analysis was performed with the software tool PhenotypeSeeker. Consequently, a list of candidate genes (mainly coding for transporters, proteins involved in the biosynthesis of cofactors, and proteins related to the metabolism of different compounds as fatty acids) was compiled. Our results suggest that in Sinorhizobium meliloti the competition abilities are linked to multiple genetic determinants that comprise several cellular components.
- Rhizobial strains with enhanced tolerance to water deficiency were selected. Rhizobial strains were isolated in Algeria directly from nodules collected from alfalfa growth in severe/moderate saline soils. The tolerance of each rhizobial strain to NaCl was determined in 96-microwell with BIOLOG technology. On the most promising isolates, single inoculation assays are performing to evaluate their nitrogen fixation efficiency. Sinorhizobium strains showing high resistance to salt stress, symbiotic efficiency, and competitiveness (detected based on genomic signatures previously identified) will be selected for the formulation of elite inoculants.
Read more
Project website: http://www.all-inproject.com/
Twitter: ALL-IN@CarloViti5
Reference paper: Bellabarba et al., 2021. Competitiveness for nodule colonization in Sinorhizobium meliloti: combined in vitro-tagged strain competition and genome-wide association analysis. mSystems
6:e00550-21. doi.org/10.1128/mSystems.00550-21
Authors
Agnese Bellabarba (Genexpress Laboratory, Department of Agronomy, Food, Environmental and Forestry (DAGRI), University of Florence, Sesto Fiorentino, Italy); agnese.bellabarba@unifi.it
Francesca Decorosi (Genexpress Laboratory, Department of Agronomy, Food, Environmental and Forestry (DAGRI), University of Florence, Sesto Fiorentino, Italy); francesca.decorosi@unifi.it
Alessio Mengoni (Department of Biology, University of Florence, Sesto Fiorentino, Italy); alessio.mengoni@unifi.it
Francesco Pini (Department of Biology, University of Bari Aldo Moro, Bari, Italy); francesco.pini@uniba.it
Abdelkader Bekki (Laboratory of Rhizobia Biotechnology and Plant Breeding -LBRAP, University Oran1, Es Senia, 31000, Algeria); bekkiabdelkader@gmail.com
Chaimaa Mecheta (Laboratory of Rhizobia Biotechnology and Plant Breeding -LBRAP, University Oran1, Es Senia, 31000, Algeria)
Sonia Sekkour (Laboratory of Rhizobia Biotechnology and Plant Breeding -LBRAP, University Oran1, Es Senia, 31000, Algeria)
Carlo Viti (Genexpress Laboratory, Department of Agronomy, Food, Environmental and Forestry (DAGRI), University of Florence, Sesto Fiorentino, Italy); carlo.viti@unifi.it
Partners
Marcello Mele (Centro di Ricerche Agro-ambientali “E. Avanzi,” University of Pisa, Via Vecchia di Masrina, 6, 56100 Pisa, Italy)
Majida Hafidi (Faculty of sciences Department of biology, University Moulay Ismail, 11201 Zitoune, 45000 Meknès, Morocco)
Khalid Azim (Institut Nationa De La Recherche Agronomique, Department of Agronomy and Machinism, INRA Av. Ennasr RP Rabat, Maroc, 415, 10000, RABAT, Morocco)
Editor: Marijke Hunninck, ILVO / Design: Christine Dilling, ICROFS
