For developers
WET for developers
This page is intended as support to WET developers, or people who are simply interested in looking into the WET code.
Downloading source codes
The download instructions below are tailored to Windows machines. For an example on how to obtain source code on Linux machines, please see FABM.net. Note: to be able to compile GOTM-WET, you must currently use the GOTM (au branch), FABM (au branch) and WET (master branch) repositories available at https://gitlab.com/wateritech-public/waterecosystemstool.
1. Downloading source codes using Git (recommended option)
To obtain the source codes of WET, GOTM and FABM using Git, you need a git client.
- First download and install the Windows version of Git itself.
- Then, for convenience, we also recommend downloading and installing the graphical git client TortoiseGit on top of Git.
After installation of these two software packages you can simply obtain the code by right-clicking in Windows Explorer within a directory where you want the source code directories, and then choose "Git Clone...". In the window that appears, set the URL, check the target directory and click OK.
Do this for the following URLs:
WET:
• URL: https://gitlab.com/wateritech-public/waterecosystemstool/wet
• suggested target directory: WET/wet-git
GOTM:
• URL: https://gitlab.com/wateritech-public/waterecosystemstool/gotm
• suggested target directory: WET/gotm-git
• Switch/Checkout to "au" branch
• do a "recursive" update of submodules (right click inside main gotm-git folder, select "TortoiseGit", then "Submodule Update...", then tick the "Recursive" box and click "Ok")
FABM:
• URL: https://gitlab.com/wateritech-public/waterecosystemstool/fabm
• suggested target directory: WET/fabm-git
• Switch/Checkout to "au" branch
To update any of the codes using Git, you simply have to right-click inside your local code repository, then select “TortoiseGit” and “Pull…". To compile the code, you need CMake and a Fortran compiler.
Please read our compilation instructions for further details.
2. Downloading source codes directly from Gitlab webpage
WET:
• To download WET directly through a website, go to https://gitlab.com/WET/wet
• Then click “Clone or download” and then “Download as zip”
• Follow the instructional video to first create a free Gitlab account and the download the source code as a zip file through the Gitlab page of WET.
GOTM:
• To download GOTM directly through a website, go to https://gitlab.com/WET/gotm
• Then click “Clone or download” and then “Download as zip”
FABM:
• To download FABM directly through a website, go to https://gitlab.com/WET/fabm
• Then click “Clone or download” and then “Download as zip”
Compilation of source codes
This guide is tailored to Windows machines, and use a build system based on CMake. For an example on how to build on Linux machines, please see FABM.net. CMake generates platform-specific build systems. In the example below, we have used CMake 3.10.0 to generate a build system for MS Visual Studio with an Intel Visual Fortran integration. You may follow the individual steps in the guide below to build a GOTM executable with FABM and WET integration.
Open CMake and point to GOTM source code directory
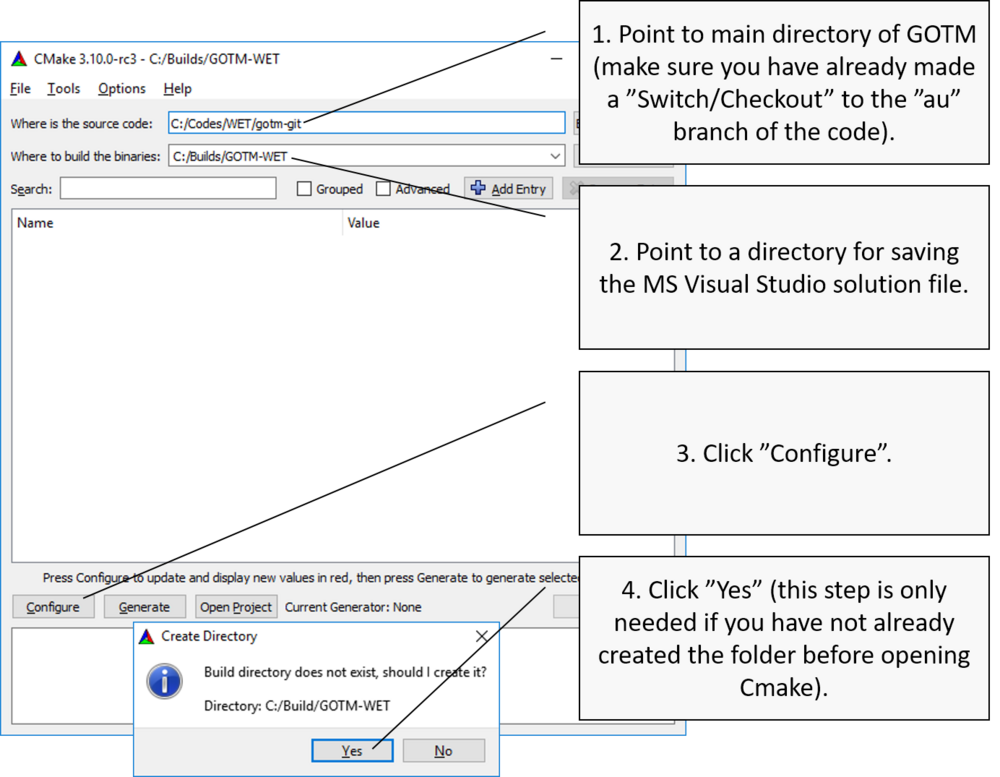
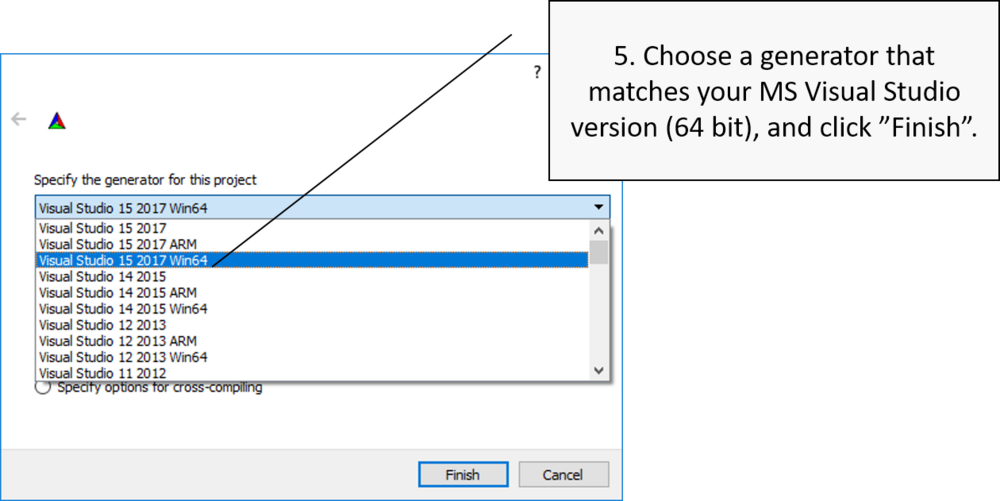
Choose build system
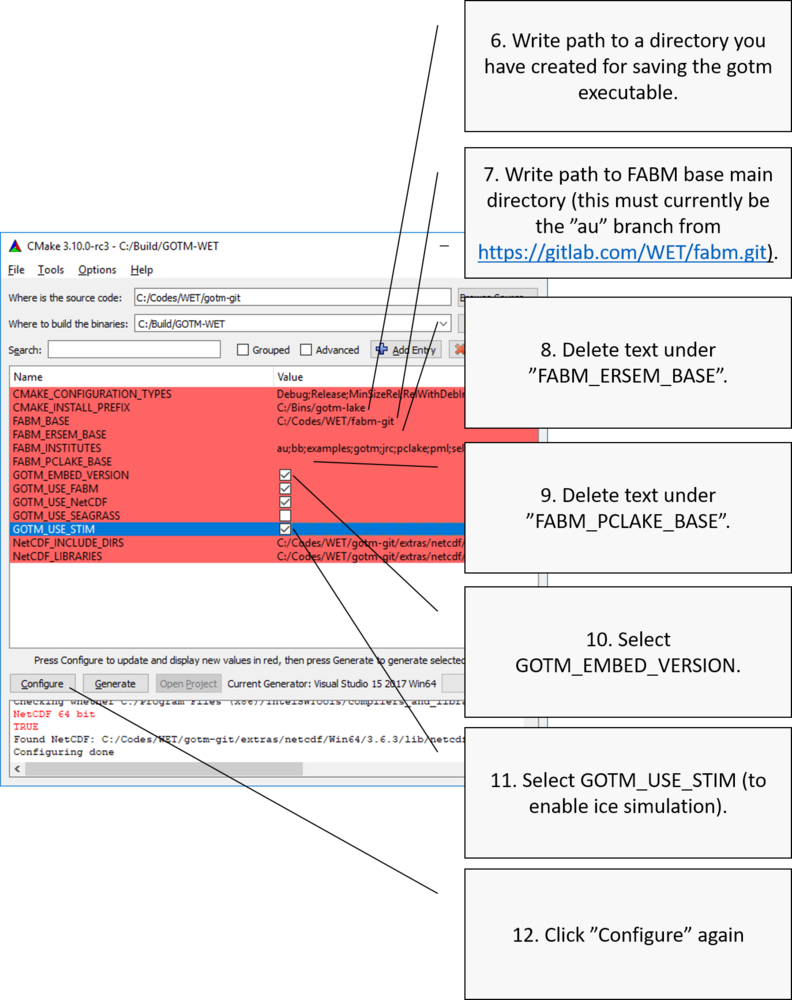
Point to FABM base
At first, you will get an error (if you get a pop-up with an error message simply click "OK"), as CMake does not know where to find the FABM code repositories (including the FABM base and WET). We need to point to FABM base, and also WET and/or ERSEM, if we want to include these in our MS Visual Studio solution.
Point to WET directory
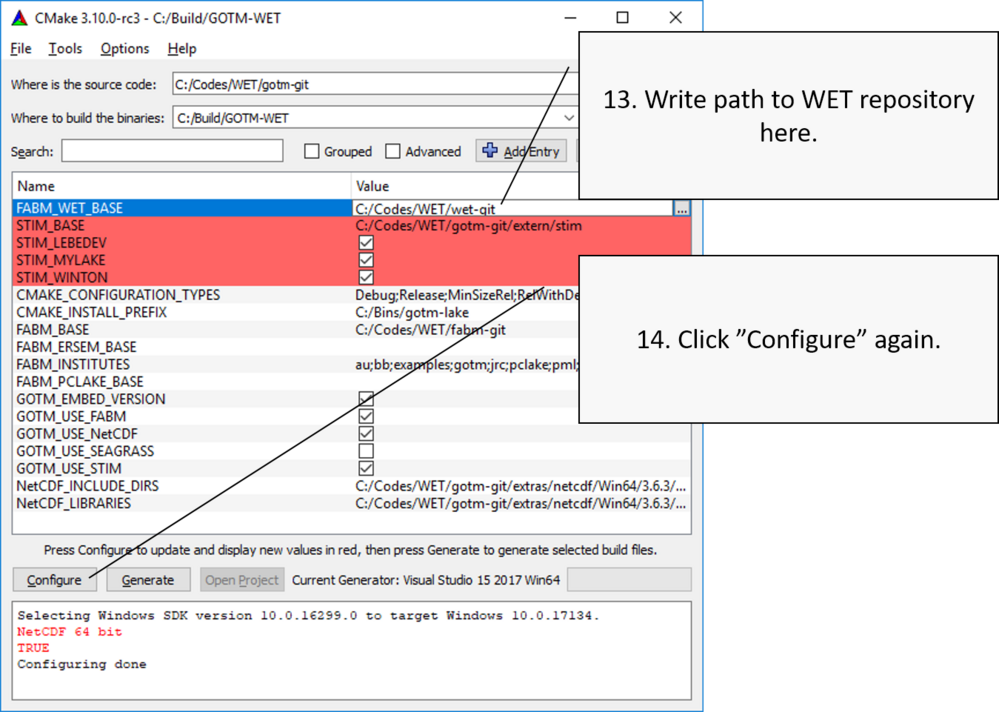
Generate MS Visual Studio solution
When Configuration is completed successfully, no menu items should be highlighted with red. Then you can create the MS Visual Studio solution file, which can be opened and used to compile the GOTM executable including FABM and the WET model. Click Generate to create this solution file.
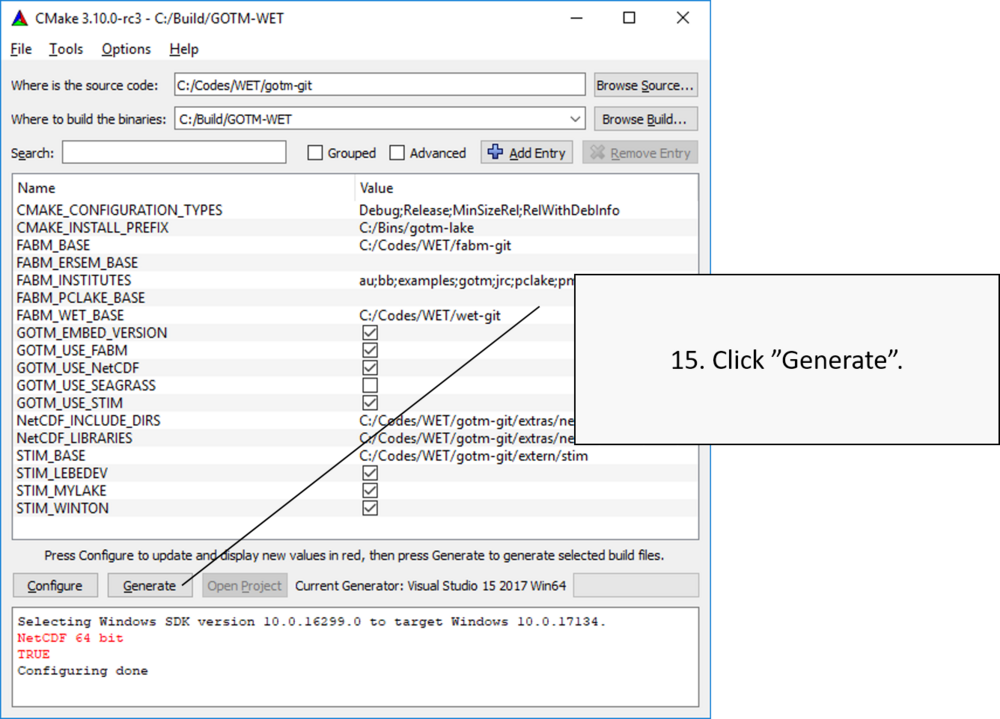
Compiling
When Generation of solution file is successful (as seen below), you can locate the MS Visual Studio solution file under your “Build” directory, which you specified in the second step of your CMake configuration (i.e. “Where to build the binaries”). You can also open the solution file by clicking on "Open Project" within CMake. You may now close CMake.
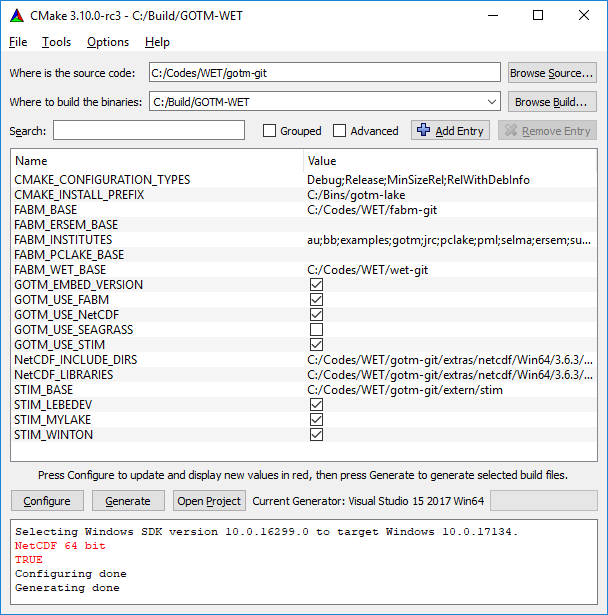
Example of build directory is seen below.
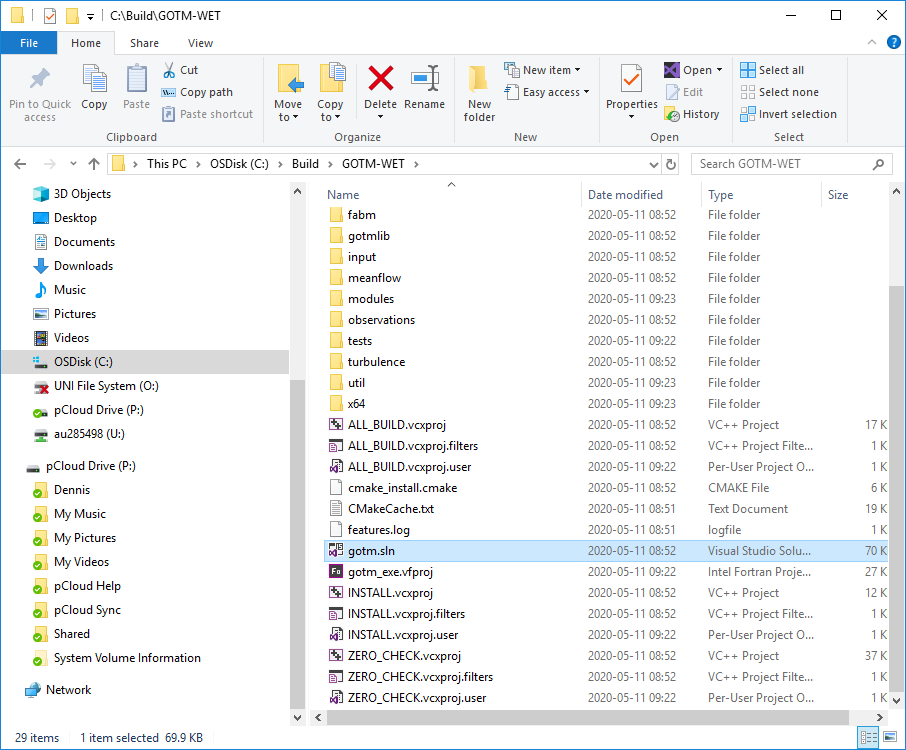
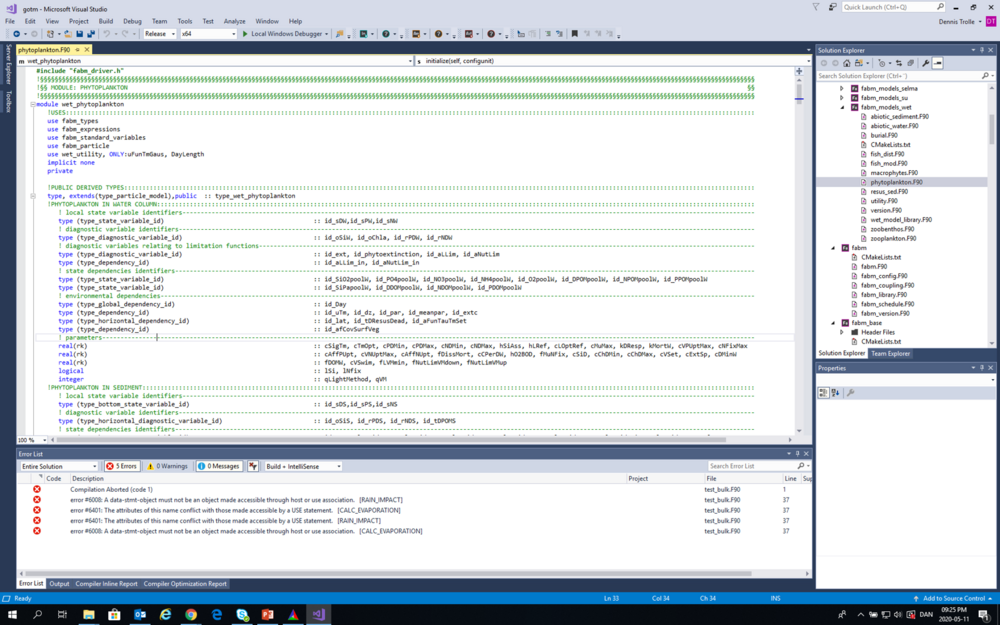
Building the GOTM executable
Open the gotm solution in MS Visual Studio. Then choose "Release" version (if you want a faster executable without debug info), then click "Build” and “Build Solution” to compile the GOTM executable with FABM and WET support. Do not be alarmed if it seems as if compilation has aborted (as illustrated below). The aborted compilation is only for an additional test module (which unfortunately cannot readily be switched off during compilation on windows machines).
You can now find the GOTM executable in a subdirectory inside the folder of your MS Visual Studio solution (e.g., in folder called Release, if you compiled in Release mode, or Debug if you compiled in Debug mode). You can also right click on the INSTALL project inside MS Visual Studio (under the Solution Explorer at your right-hand side) and click “Build” – this will copy the gotm executable to the directory you have specified in your CMake configuration.
Modifying WET source code
Within the GOTM solution, when open in MS Visual Studio, you can find the WET source files under the Solution Explorer (your right-hand side) under fabm\models\fabm_models_wet\. You can review and change code within these files, and then re-build the gotm executable. Please see FABM.net for further details on how to write FABM compliant code for a biogeochemical model.



