Quick Wins
Quick wins
-
Developing a Standardized Optimized Soil Sampling Procedure for Microbial and Physico-chemical Analyses
-
Sampling Biodiversity across Europe
-
Developing a Standardized Optimized Procedure for Microbial DNA Extraction from Soils
-
Soil protozoa: Abundant, diverse, but largely unknown organisms
-
Earthworms are not all the same - even in the same functional Group
-
Developing and Standardising Tools and Procedures for Assessment of Soil Biodiversity
Developing a Standardized Optimized Soil Sampling Procedure for Microbial and Physico-chemical Analyses
Soil is known to be a naturally complex and heterogeneous environment, both in space and time, and both from a physico-chemical and from a biological point of view. Soil microbial communities have indeed been shown to vary at different scales, from the millimeter to the continent. Therefore, the soil sampling procedure is of crucial importance for studying soil microbial communities, especially in large scale studies for which several teams of samplers are involved. This is the reason why a Standardized Optimized soil sampling Procedure (SOP) for microbial and physico-chemical analyses has been developed by the GenoSol platform within the EcoFINDERS program, and spread to all the partners involved in soil sampling.

The SOP describes:
- the materials needed to perform soil sampling.
- the soil sampling procedure itself, specially designed to obtain soil samples representative of a designated area.
- the guidelines to properly homogenize and pack samples for long term storage.
The SOP has been made available to all the EcoFINDERS partners through an online collaborative platform, and has been illustrated by a short online demonstration video publicly available at:
http://www.youtube.com/watch?v=_k7BEInBXEc&feature=youtu.be
Thanks to the dissemination of such an SOP, large scale sampling event during fall 2012 at several European sites was possible (see quick win b), thus demonstrating the feasibility of reproducible large scale samplings.
For further information and updates contact:
Pierre Plassart (pierre.plassart@dijon.inra.fr)
Sampling biodiversity across Europe
In September 2012 an unprecedented sampling event took place. Instigated by the need to establish the NormalOperatingRange (or NOR) of soil biodiversity indicators for policy development, the EcoFINDERS project undertook a European wide campaign to sample soil biodiversity and ecosystem function across differing land uses and climatic zones. This huge sampling event, co-ordinated by Teagasc, involved 10 partner organisations and the sampling of 81 sites to deliver a snap-shot of biodiversity in European soils.
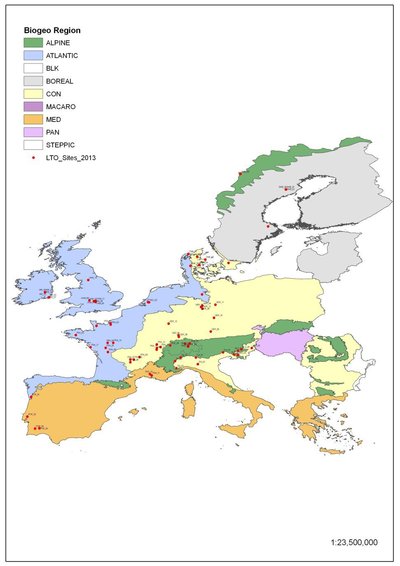
Figure 1. Distribution of 81 sites representing a pedo-climatic transect across Europe.
Intensive planning was needed in order to ensure the smooth running of the sampling campaign. A coding system was developed to give each sample a unique identifier, whilst at the same time a database design was invented for long-term data storage of the results collected. Detailed SOPs (with step by step instructions and photographs to ensure clarity despite different language speakers) were designed and sent out to partners. To ensure that all samples were identical, 2000 plastic cores were prepared by Teagasc and couriered to those doing the sampling inside sample packs containing everything the scientist in the field might possibly need.

Picture 1 & 2. Sample packs contained cores for sampling, field cards, string for measuring out the sample area and a detailed SOP.
Throughout September, October and November, sample packs and soils flew back and forth over Europe, as scientists went out in all weathers (Slovenia even sampled in the snow) to hammer cores into the ground and send back the valuable samples.


| Picture 3. Sampling in Slovenia, in the snow. | Picture 4. Packs were returned to Teagasc in insulated boxes, shipped with freezer packs to keep the samples cool. |
In the laboratories of Teagasc, a small team worked throughout the early winter, racing to get fresh soils sieved, sub-sampled and back out to partners for analysis as quickly as possible. A secondary aim of the Transect was to prove that the time-dependant nature of many of the parameters measured did not have to be a barrier to a wide-spread sampling campaign and we are pleased to report that this was successfully achieved.
Analysis is currently being carried out in labs all over Europe, with results already being collected by the data manager at Teagasc. The full dataset is expected to be complete by the end of 2013, though results are already being shared between partners through the Collaborative Platform (hosted by INRA).
Table 1. Biological indicators applied in Long- Term Observatories and Transect sites across Europe.
Biodiversity | Function | LTO | Transect |
TRFLP* (DNA&) |
| X | X |
Protozoa (DNA) |
| X | X |
PLFA** |
| X | X |
Fungi (ergosterol) |
| X | |
FG*** nitrification (DNA) | X | ||
FG denitrification (DNA) | X | ||
Earthworms | X | ||
Enchytraeids | X | X | |
Micro-arthropods | X | X | |
Nematodes | X | X | |
| Bait Lamina | X | |
| Water infiltration | X | |
| Resilience | X | |
| Nitrification | X | X |
| HW-C & PM-N**** | X | |
| Micro-resp | X | X |
| Biolog | X | |
| Enzyme Activity | X | X |
| FG Supressiveness (DNA) | X |
|
- *Terminal Restriction Fragment Length Polymorphism
- & analysis of DNA extracted from soil
- **Phospho-lipid fatty-acid analysis
- ***Functional gene analysis
- ****Hot water extractable carbon and potentially mineralisable nitrogen
The final dataset will be unique and all partners involved are looking forward with great excitement to exploring the new ecological information uncovered.
Dote Stone and Rachel Creamer
Developing a Standardized Optimized Procedure for Microbial DNA Extraction from Soils
Soil DNA extraction has always been a critical step in describing microbial biodiversity. Historically, ascertaining overarching microbial ecological theories has been hindered as independent studies have used numerous custom and commercial DNA extraction procedures. For that reason, a standardized soil DNA extraction method (ISO-11063) was recently published. However, although this ISO standard is suited for molecular tools such as quantitative PCR and community fingerprinting techniques, it has only been optimized for examining soil bacteria. Therefore, an appropriate soil DNA extraction procedure for examining bacterial, archaeal and fungal diversity in soils of contrasting land use and physico-chemical properties was developed by the GenoSol platform.
This new DNA extraction procedure is an optimization of the ISO-11063 standard and has therefore been named ISOm (for Modified ISO). The modification of the ISO standard lies in the first step of the DNA extraction, which is a physical grinding of soil. Indeed, the ISO standard recommends mixing soil with glass beads of different sizes and shaking it in a one dimension shaker, this resulting in breaking bacterial cells. The ISOm procedure uses a different set of glass beads to which silica and ceramic beads are added, and recommends using a three dimension shaker. This grinding optimization results in breaking tougher cell walls. It has thus been demonstrated that the ISOm DNA extraction procedure yields more total DNA from soil, more bacterial DNA, more archaeal DNA, and more fungal DNA than the ISO standard.
The ISOm DNA extraction procedure was published in 2012 (Plassart P, Terrat S, Thomson B, Griffiths R, Dequiedt S, Lelièvre M, Regnier T, Nowak V, Bailey M, Lemanceau P, Bispo A, Chabbi A, Maron PA, Mougel C, Ranjard L (2012) Evaluation of the ISO standard 11063 DNA extraction procedure for assessing soil microbial abundance and community structure. PLoS ONE 7(9): e44279. doi:10.1371/journal.pone.0044279).

In this paper, it was shown that the choice of DNA extraction method can have a significant effect upon bacterial, archaeal and fungal molecular analyses and is therefore an important consideration for microbial studies. This is particularly the case for soil fungi as increased fungal abundances were detected using the ISOm procedure, and extraction protocol was generally found to have more effect upon fungal community structure than soil type. Specifically, the effects upon community structure were less pronounced using the ISO standard compared to the ISOm procedure. Greater yields of DNA and increased abundances were also measured with the ISOm procedure. This new soil microbial DNA extraction procedure therefore seemed to be a great tool for the EcoFINDERS project. Indeed, all the EcoFINDERS partners studying soil microbial communities, whether focusing on bacteria, archaea, or fungi, can use the exact same DNA sample and consequently share and compare their results in an easier way.
The ISOm soil microbial DNA extraction procedure has been applied to more than 450 samples resulting from large scale samplings that happened throughout Europe for the EcoFINDERS project (see quick win b).
For more information and updates contact:
Pierre Plassart (pierre.plassart@dijon.inra.fr)
Soil protozoa: Abundant, diverse, but largely unknown organisms
Stefan Geisen1, Michael Bonkowski1
1Zoological Institute, Dept. Terrestrial Ecology, University of Cologne, Germany
Protozoa occupy key positions in soil food webs; they are the major consumers of bacterialproduction in soil due to their small size, high abundance and fast turnover rates. Despite their important functions for nutrient cycling at the base of the heterotrophic eukaryotic food web in soil we only have a vague idea on the identity of the dominant taxa. This lack of knowledge is caused by widespread methodological difficulties in cultivation and quantification in the opaque soil environment as well as a lack of taxonomic expertise. Especially naked amoebae have received little attention in the past, due to difficulties both, in isolation and identification. However, recent developments in molecular techniques now promise to reduce this methodological gap on this largely unknown trophic link in the soil food web.
We combined morphological and molecular methods to determine the normal operating range on the occurrence of soil naked amoebae within the EU-project EcoFINDERS.
Quick win of EcoFINDERS grant agreement No 264465
Earthworms are not all the same - even in the same functional group

If there was a contest on which animal group contributes the most to soil ecosystem functioning, earthworms might take the top spot. Decomposition, soil fertility, plant growth, soil structure, microbial activity, organic matter turnover and stabilization, greenhouse gas emissions: there’s hardly a function in which they don’t play a role. However, not all earthworms are the same: different species do different things. Some prefer to feed on plant litter and dung, while others literally eat soil. Some dig extensively in the soil (they are “nature’s plough”, as Charles Darwin said), others make few but large and enduring channels, and yet others mostly live above the soil, for instance beneath leaf litter.
To make sense of this diversity, scientists often use functional groups to combine species according to their characteristics. Species in the same group are supposed to perform the same function in a similar way. Anecic earthworms are especially important as “ecosystem engineers”: they incorporate large amounts of organic detritus inside the burrows they make, semi-vertical and open at the surface, which also enhance water flow in the soil and are hotspots of biological activity.
But are different species functionally redundant, that is, do they have the same effects?

Field site at LTO Lusignan, France
To investigate this, in spring 2013 we performed a field experiment in a grassland field of the Lusignan LTO (France), where we compared the effects of two anecic earthworm species, Aporrectodea longa and Lumbricus centralis. Using a method we had tested in a previous study, we placed plant litter enriched in stable isotopes (13C and 15N) around the opening of their burrows to track the incorporation of C and N in the soil.
 Earthworm cast next to burrow entrance
Earthworm cast next to burrow entrance
After 50 days we sampled the soil around the burrows and found that L. centralis had incorporated more 13C, and thus organic matter from the plant litter, than A. longa. This was also reflected in the biochemistry of the soil organic matter, analysed in collaboration with external partners Cornelia Rumpel and Phuong Ngo (BIOEMCO, France): with L. centralis there was a higher contribution of fresh, plant-derived sugars, while with A. longa (as well as without earthworm) there were relatively more microbial-derived, highly decomposed sugars. Overall, the soil around L. centralis burrows was biochemically different than soil around A. longa burrows.
In short, we found that even in the same functional group earthworm species are not all the same.
Another demonstration from EcoFINDERS on the importance of soil biodiversity and the need to study it more closely.

Earthworm burrow entrance
For further information and updates, contact:
Walter Andriuzzi (walter.andriuzzi@wur.nl)
Read more (the previous study where we tested the methodology):
Andriuzzi, W.S., Bolger, T, Schmidt, O. The drilosphere concept: Fine-scale incorporation of surface residue-derived N and C around natural Lumbricus terrestris burrows. Soil Biology & Biochemistry. Read the article
Developing and Standardising Tools and Procedures for Assessment of Soil Biodiversity
Emilie Tisserand, Pierre Plassart, Marc Buée, Juliette Lengelle, Thibaud Payen, Claude Murat, Stéphane Uroz, Philippe Lemanceau, and Francis Martin
Soil is the reservoir of a high diversity of organisms. This biodiversity plays an important role in ecosystem functioning and nutrient cycling due to the ability of these organisms to degrade organic matter, weather minerals, and fix carbon and nitrogen. However, we are far from appreciating this telluric black box of diversity. Most of our present knowledge was established using cultivation-dependent approaches giving access to only a limited portion of this diversity. The development in the last decade of high-density 16S rRNA, 18S rRNA or ITS microarrays, monogenic and shotgun metagenomics has permitted us to greatly improve our knowledge, generating very informative views on the structure and diversity of the soil microbiome.
In the frame of the EcoFINDERS project, the impact of the type and intensity of land management was tested across Europe in different long-term observatories using amplicon-based metagenomics. The objectives were to decipher the diversity and structure of a wide range of organisms including archaea, bacteria, fungi (Glomeromycota and Dikarya), protists, and nematodes. This approach was a scientific challenge because our present knowledge of the diversity and associated molecular markers of the range of organisms are not at the same level. Here, we present the tools to investigate soil biodiversity and we discuss the major results highlighting the diversity of archaea, bacteria, fungi, amoebozoa, flagellates and nematodes in European soils.
For most of the organisms considered ribosomal genes have been used as marker genes: i) 16S rRNA for archea and bacteria and 18S rRNA for the others. The internal transcribed spacer region (region ITS1) was used to decipher the fungal diversity, focusing on Dikarya sub-kingdom (Basidiomycota and Ascomycota). The same metagenomic DNA samples were used to assess to the diversity of all the soil organisms. After amplification, the sequencing of these marker genes generated a data set containing from 245,867 sequences for the amobozoa to 3 millions of sequences for the bacteria. The next step was to analyse this large diversity of sequences. To date, several bioinformatic pipelines have been developed such as RDP (ribosomal data project, Cole et al., 2009), Mothur (Schloss et al., 2009), QIIME (Caporaso et al., 2010) and UPARSE (Edgar, 2013), and most of them have been optimized to analyse the bacterial diversity. As one important task of the project was to analyse the sequences generated for each type of organism in a similar way, different pipelines have been tested and one composite pipeline - named pipeline #1 - described in Figure 1, was at last established to analyse all the sequences.