Environmental DNA (eDNA) is defined as genetic material obtained directly from environmental samples such as soil, sediment, water, snow, etc. or leftovers from an organisms such as hair, feces or carcasses. eDNA is an efficient, non-invasive and easy-to-standardize sampling approach with great potential as a monitoring tool for wildlife. It constitutes a great opportunity to study the role and ecology of organisms and get insight into their metabolism, function and evolutionary relationship.
Studying eDNA involves a series of methods based on DNA collected directly from the environment to identify the presence of organisms either by Q-PCR or by metabarcoding at the individual, population, species, genus or family level or for the determination of biodiversity and ecosystem characterization. Both within the terrestrial and aquatic environment, it is expected that eDNA will play a significant role in future environmental monitoring. eDNA builds on skills in molecular biology, population genetics and bioinformatics coupled with ecology, biology and/or agronomy.
The fast development within the area of DNA and RNA sequencing techniques combined with the reduction in prices of sequencing analysis have strengthened the interest in wide-spread use of techniques based on eDNA for monitoring and research applications.
In the eDNA center at Aarhus University, Faculty of Technical Sciences investigations of potentials and feasability of employing eDNA in environmental monitoring of both state of environment and tracking of specific organisms will performed. The objective is to perform preparatory investigations facilitating incorporation of eDNA techniques in environmental monitoring.
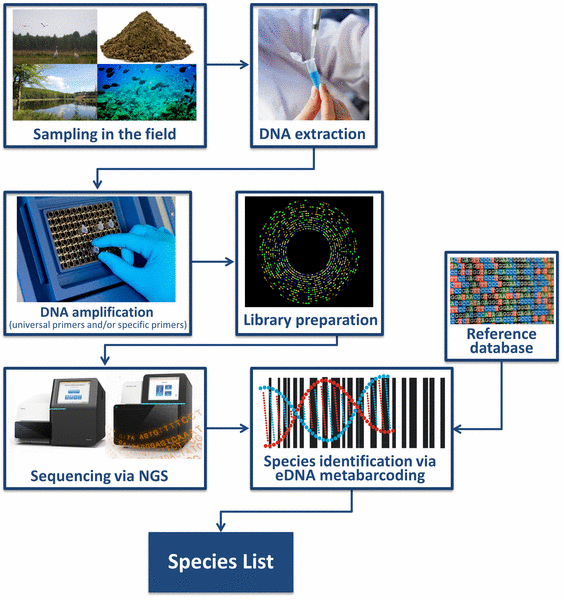
Analysis of eDNA involves the collection of a small volume of water, soil, sediment, feces, etc which is sent to the laboratory where it is treated to extract the eDNA present. This method uses either universal primers that amplify DNA from a group of target species (e.g. bacteria, amphibians, fishes, crustaceans, plants, etc.) or total environmental DNA as input. After amplification in the PCR process or shotgun library preparation the DNA fragments are subsequently sequenced using in-house state of the arts sequencing platforms. The resulting sequences are then assembled to genes/genomes or compared with a reference database to establish a list of species from which DNA is present in the sample. Accordingly, eDNA is used to address applied and fundamental research questions within areas ranging from molecular biology, ecology, palaeontology and environmental sciences. ![]()